The Qian Lab
California Institute of Technology
Experimental work
[17] Heat-rechargeable computation in DNA logic circuits and neural networks. Tianqi Song and Lulu Qian. Nature, 10.1038/s41586-025-09570-2, 2025. (pdf) (supp info) (source data and code)

[16] Supervised learning in DNA neural networks. Kevin M. Cherry and Lulu Qian. Nature, 645:639–647, 2025. (pdf) (supp info) (source data and code)

[15] Modular reconfiguration of DNA origami assemblies using tile displacement. Namita Sarraf, Kellen R. Rodriguez, and Lulu Qian. Science Robotics, 8:eadf1511, 2023. (pdf) (supp info) (DNA29 slides)

Focus: Reconfigurable self-assembled DNA devices. Erik Benson and Jonathan Bath. Science Robotics, 8:eadh8148, 2023.
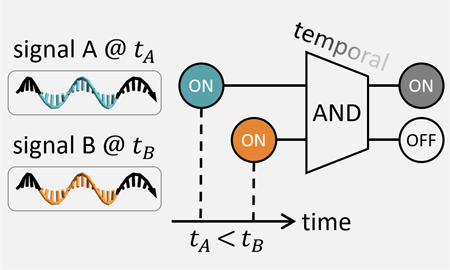
[14] DNA strand-displacement temporal logic circuits. Anna P. Lapteva, Namita Sarraf, and Lulu Qian. JACS, 144:12443−12449, 2022. (pdf) (supp info)
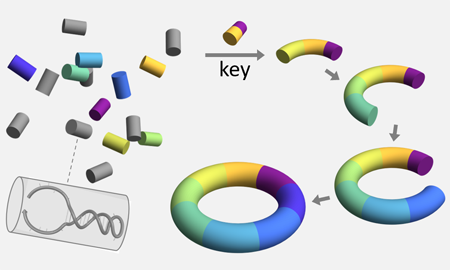
[13] Developmental self-assembly of a DNA ring with stimulus-responsive size and growth direction. Allison T. Glynn, Samuel R. Davidson, and Lulu Qian. JACS, 144:10075−10079, 2022. (pdf) (supp info)
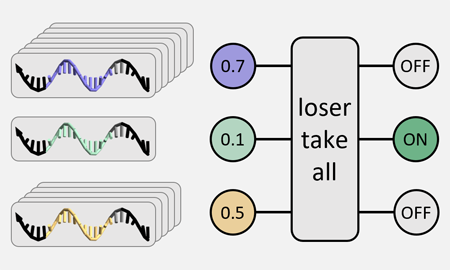
[12] A loser-take-all DNA circuit. Kellen R. Rodriguez, Namita Sarraf, and Lulu Qian. ACS Synthetic Biology, 10:2878−2885, 2021. (pdf) (supp info)
[11] A cooperative DNA catalyst. Dallas N. Taylor, Samuel R. Davidson, and Lulu Qian. JACS, 143:15567−15571, 2021. (pdf) (supp info)
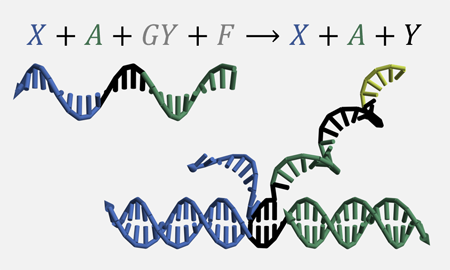
[10] Information-based autonomous reconfiguration in systems of interacting DNA nanostructures. Philip Petersen, Grigory Tikhomirov, and Lulu Qian. Nature Communications, 9:5362, 2018. (pdf) (supp info) (supp movie)
[9] Triangular DNA Origami Tilings. Grigory Tikhomirov, Philip Petersen, and Lulu Qian. JACS, 140:17361−17364, 2018. (pdf) (supp info)
[8] Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Kevin M. Cherry and Lulu Qian. Nature, 559:370–376, 2018. (pdf)

Caltech news story Media coverage
[7] Probabilistic switching circuits in DNA. Daniel Wilhelm, Jehoshua Bruck, and Lulu Qian. PNAS, 115:903–908, 2018. (pdf) (supp info)
[6] Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Grigory Tikhomirov, Philip Petersen, and Lulu Qian. Nature, 552:67–71, 2017. (pdf) (supp info)

News and Views: DNA self-assembly scaled up. Fei Zhang and Hao Yan. Nature, 552:34–35, 2017.
Caltech news story Media coverage
[5] A cargo-sorting DNA robot. Anupama J. Thubagere, Wei Li, Robert F. Johnson, Zibo Chen, Shayan Doroudi, Yae Lim Lee, Gregory Izatt, Sarah Wittman, Niranjan Srinivas, Damien Woods, Erik Winfree, and Lulu Qian. Science, 357:eaan6558, 2017. (pdf) (supp info)
Perspective: DNA robots sort as they walk. John H. Reif. Science, 357:1095–1096, 2017.
Caltech news story Media coverage
[4] Compiler-aided systematic construction of large-scale DNA strand displacement circuits using unpurified components. Anupama J. Thubagere, Chris Thachuk, Joseph Berleant, Robert F. Johnson, Diana A. Ardelean, Kevin M. Cherry, and Lulu Qian. Nature Communications, 8:14373, 2017. (pdf) (supp info)
[3] Programmable disorder in random DNA tilings. Grigory Tikhomirov, Philip Petersen, and Lulu Qian. Nature Nanotechnology, 12:251–259, 2017. (pdf) (supp info)

In the Classroom: Programming random mazes. Philip Petersen. Nature Nanotechnology, 12:284, 2017. (pdf)
News and Views: DNA origami tiles: Nanoscale mazes. Fei Zhang, Fan Hong and Hao Yan. Nature Nanotechnology, 12:189–190, 2017. (pdf)
[2] Neural network computation with DNA strand displacement cascades. Lulu Qian, Erik Winfree, and Jehoshua Bruck. Nature, 475:368–372, 2011. (pdf) (supp info)
News and Views: DNA and the brain. Anne Condon. Nature, 475:304–305, 2011. (pdf)
[1] Scaling up digital circuit computation with DNA strand displacement cascades. Lulu Qian and Erik Winfree. Science, 332:1196–1201, 2011. (pdf) (supp info)
Perspective: Scaling up DNA computation. John H. Reif. Science, 332:1156–1167, 2011. (pdf)
News and Views: DNA computes a square root. Yaakov Benenson. Nature Nanotechnology, 6:465–467, 2011. (pdf)
Theoretical work
[6] Two-dimensional tile displacement can simulate cellular automata. Erik Winfree and Lulu Qian. arXiv, arXiv:2301.01929, 2023. (pdf)

[5] Simplifying chemical reaction network implementations with two-stranded DNA building blocks. Robert F. Johnson and Lulu Qian. In C. Geary and M. J. Patitz, editors, DNA Computing and Molecular Programming, LIPIcs, 174:2:1–2:14, 2020. (pdf)
[4] Programming and simulating chemical reaction networks on a surface. Samuel Clamons, Lulu Qian, and Erik Winfree. Journal of the Royal Society Interface, 17:20190790, 2020. (pdf)
[3] Parallel and scalable computation and spatial dynamics with DNA-based chemical reaction networks on a surface. Lulu Qian and Erik Winfree. In S. Murata and S. Kobayashi, editors, DNA Computing and Molecular Programming, LNCS, 8727:114–131, 2014. (pdf)
[2] Efficient Turing-universal computation with DNA polymers. Lulu Qian, David Soloveichik, and Erik Winfree. In Y. Sakakibara and Y. Mi, editors, DNA Computing and Molecular Programming, LNCS, 6518:123–140, 2011. (pdf)
[1] A simple DNA gate motif for synthesizing large-scale circuits. Lulu Qian and Erik Winfree. Journal of the Royal Society Interface, 8:1281–1297, 2011. (pdf) Conference version in A. Goel, F. C. Simmel and P. Sosik, editors, DNA Computing and Molecular Programming, LNCS, 5347:70–89, 2009. (pdf)